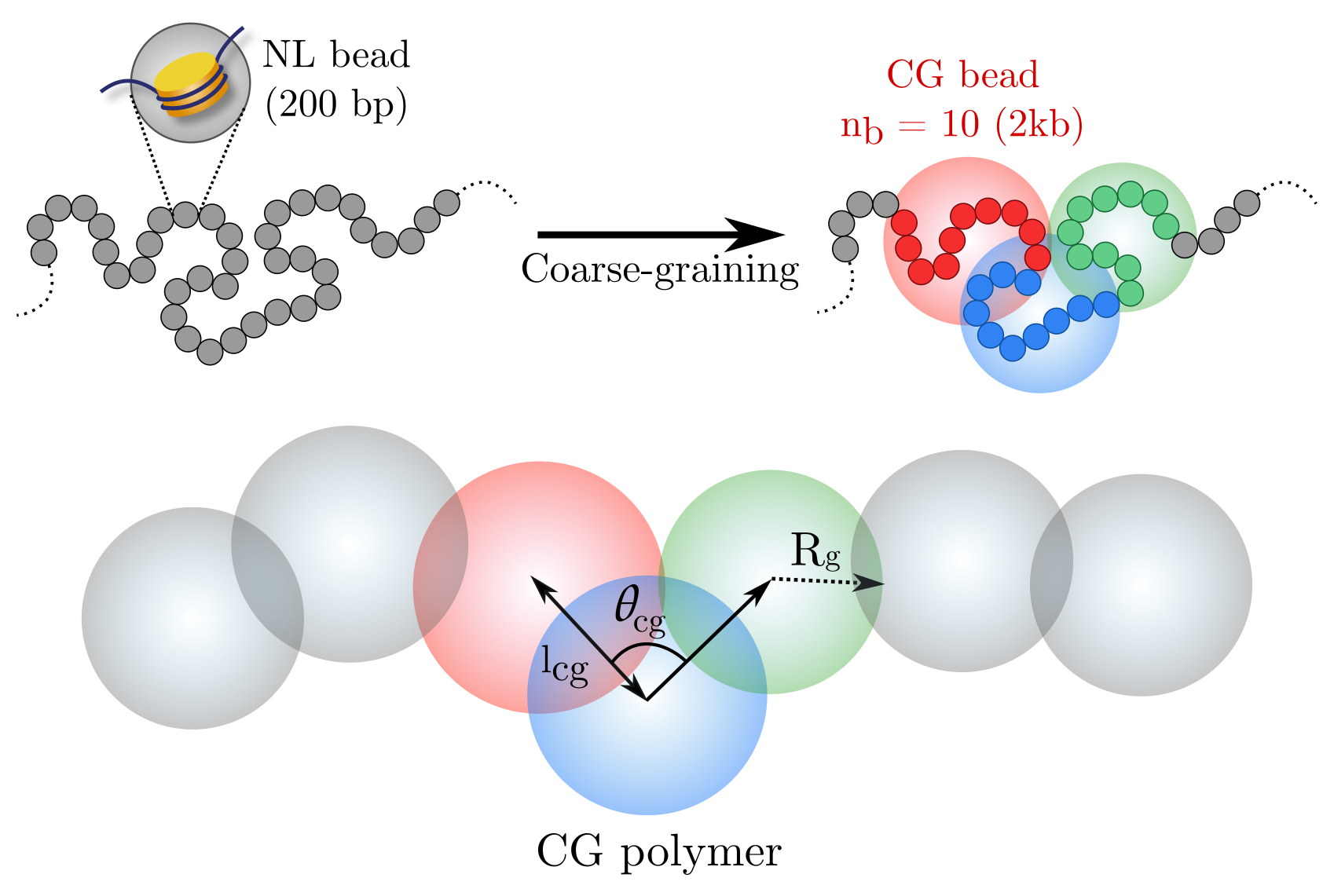
Predicting Scale-Dependent Chromatin Polymer Properties from Systematic Coarse-Graining
The organization and dynamics of chromatin are essential for understanding gene regulation and cell function. In order to simulate these processes accurately, researchers use coarse-grained bead-spring polymer models to describe chromatin. However, the dimensions of the beads, their elastic properties, and the nature of the inter-bead potentials have been largely unknown.
To address this, we utilized publicly available nucleosome-resolution contact probability (Micro-C) data to systematically coarse-grain chromatin and predict important quantities for polymer representation of chromatin. We computed size distributions of chromatin beads for different coarse-graining scales, quantified fluctuations and distributions of bond lengths between neighboring regions, and derived effective spring constant values.
Our findings suggest that contrary to prevalent notions, coarse-grained chromatin beads should be considered soft particles that can overlap. We derived an effective inter-bead soft potential and quantified an overlap parameter, which provides crucial insights into the nature of chromatin packing. We also computed angle distributions between neighboring bonds, which gave insights into intrinsic folding and local bendability of chromatin.
Interestingly, we found that the bead sizes, bond lengths, and bond angles showed different mean behavior at the boundaries of chromatin domains. We integrated our findings into a coarse-grained polymer model and provided quantitative estimates of all model parameters, which can serve as a foundational basis for all future coarse-grained chromatin simulations.
The organization and dynamics of chromatin are essential for understanding gene regulation and cell function. In order to simulate these processes accurately, researchers use coarse-grained bead-spring polymer models to describe chromatin. However, the dimensions of the beads, their elastic properties, and the nature of the inter-bead potentials have been largely unknown.
To address this, we utilized publicly available nucleosome-resolution contact probability (Micro-C) data to systematically coarse-grain chromatin and predict important quantities for polymer representation of chromatin. We computed size distributions of chromatin beads for different coarse-graining scales, quantified fluctuations and distributions of bond lengths between neighboring regions, and derived effective spring constant values.
Our findings suggest that contrary to prevalent notions, coarse-grained chromatin beads should be considered soft particles that can overlap. We derived an effective inter-bead soft potential and quantified an overlap parameter, which provides crucial insights into the nature of chromatin packing. We also computed angle distributions between neighboring bonds, which gave insights into intrinsic folding and local bendability of chromatin.
Interestingly, we found that the bead sizes, bond lengths, and bond angles showed different mean behavior at the boundaries of chromatin domains. We integrated our findings into a coarse-grained polymer model and provided quantitative estimates of all model parameters, which can serve as a foundational basis for all future coarse-grained chromatin simulations.